Mathematical Models for Cellular Destination Prediction
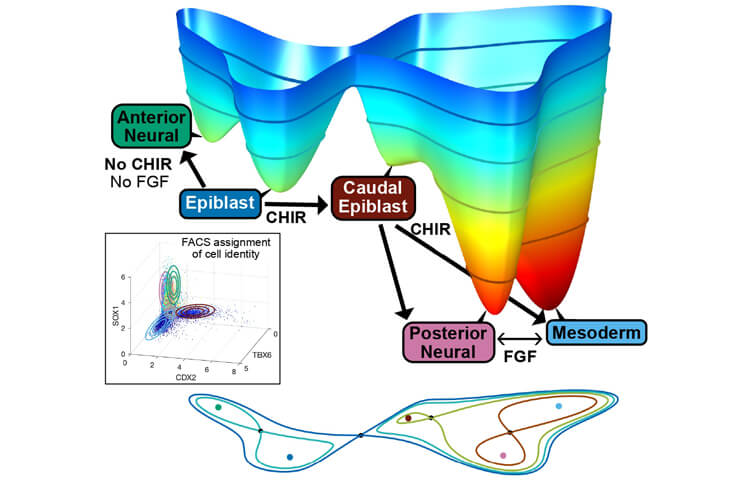
The generation of cellular diversity during the development of an embryo into an adult involves the differentiation of cells between discrete cellular states. In the 1940s, developmental biologist Conrad Hal Waddington introduced the metaphor of a landscape to describe this process, which explains the development of a cell as a ball rolling through valleys, corresponding to different cell types.
Based on this model, Dr Meritxell Sáez with the Department of Quantitative Methods at the IQS School of Engineering is conducting a line of research on “Mathematical Modelling of Embryonic Development Biology” alongside Professor David Rand with the University of Warwick and Dr James Briscoe with the Francis Crick Institute in London. The researchers have succeeded in converting the Waddington metaphor into a prediction model of cellular evolution.
Mathematical modelling of embryonic development biology
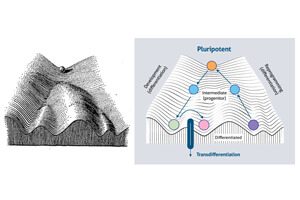
According to the model proposed by Waddington, a cell advances in its development by crossing a series of points that force it to “make decisions” about where it wants to evolve, becoming more and more specialized.
The mathematical model developed by Dr Sáez and her team seeks to understand certain events at the molecular level in response to various stimuli, without going to the mechanical detail and only observing the events that occur. The aim is to be able to predict new events in response to modifications of the stimuli.
Therefore, it represents a predictive and dynamic mathematical model with a more sophisticated “landscape” that can predict cell development through different saddle points on the graph. It reproduces the fact that the cell has become part of one tissue or another, for example, predicting that a specific cell can end up forming part of the spinal cord system or bone tissue. This will depend on the “well” (attractor) of the graph in which this cell ends, a fact that simultaneously depends on the intracellular events and signals that it has received.
This complex model is created with data from “in vitro” studies regarding single cells, which give expression information of some proteins in different cell types, seeking to find a “landscape map” system that is able to reproduce it with appropriate parameters that depend on the signals.
For the time being, this mathematical model only aims to predict cell behaviour and understand it in depth. In the future, specific applications could be sought in certain medical areas, such as mutation studies or cellular reprogramming.
Related publications
M. Saez, J. Briscoe, D.A. Rand, Dynamical landscapes of cell fate decisions, Interface Focus, 12, June 2022

